Most of use know about chain letters, specifically the financial pyramid ones where you send X dollars to the guy at the top of the addressee list as instructed, and then you make and send Y copies of the letter with the top addressee dropped, the others moved up, and you added at the bottom. After waiting a short period of time, you should receive a fortune of Z dollars.
Yes, they do work if you have an infinite population and you are able to select people at each round who have not played before (and will all play). So, in what ways does genetic genealogy appear to be like chain letters? Just as people often are added to more than one recipient’s list of people to get the letter in the future, we can come back to the same person as a common ancestor via more than one of our lines. The implicit assumption is that we have an infinite population with chain letters, but the same seems to hold for genealogy. Some will make statements to acknowledge the problem in genealogy with multiple lines from an individual going back to same ancestor, but then will proceed full speed with a methodology which ignores what they said. But the problem is even worse than with chain letters. The current world population is over 7.5 billion, so ignoring issues of individuals’ age, that is our pool of potential players. With genealogy, each generation goes back further back in time. The potential players become a lot less the further back one goes. The current population in excess of 7.5 billion grew from a level of 6 billion in the year 2000. But go 200 years further back to 1800 and the population level was only passing a billion people. So just as with chain letters where many people are asked to play more than once, negating their potential gains, we find common ancestors who played more than once in many people’s family trees.
What does this mean for our matching methodology with genetic genealogy? I assume that people who have found their way to this website probably have taken an autosomal DNA test such as FTDNA’s Family Finder or AncestryDNA and have at least a basic knowledge about how DNA works. They may be frustrated with the faults of the current matching methodology, at least in finding ancestors more than a few generations back. If you are instead a novice in need of education, plenty of information and links can be found at the International Society of Genetic Genealogy (ISOGG) Wiki site:
ISOGG Wiki
Though I have referred you to this body of knowledge, in my upcoming blog posts, I will be contradicting some of the theory contained there.
I trust that everyone at this point knows about the twenty-two pairs of autosomal chromosomes found in the nucleus of all our cells, with one chromosome in each pair being a copy of one originally supplied by a parent. We will save the 23rd chromosome pair, the X-Y sex determining one for later. Also, readers should understand the recombination process in which each parent passes down a single chromosome to a child. This chromosome has parts from the two which came from the parent’s parents. In this process the child gets half of its DNA from each parent. A chromosome passed on by a parent should average half from each of the parent’s parents’ DNA, but because of the random nature of recombination the proportion may vary a bit as one parent may contribute more and the other less. In general terms, for each additional generation, the amount of an ancestor’s DNA passed on should fall by about a half on average. Thus, a grandparent may contribute a fourth, a great grandparent an eighth, and so on until none is left. However, as we see by the Chain Letter Fallacy, an ancestor’s contribution may last further forward by having more than one lines from him to the descendant.
To determine whether two people are related, we compare their DNA results (kits) to look for sequences of DNA which are the same and presumably come from a common ancestor. The different DNA vendors have their own matching criteria and you have sites such as GEDmatch.com which have matching utilities independent from the vendors where users can upload their kits to widen their search from the vendor’s database. To be considered a likely match, a segment must exceed a set minimum length (usually expressed in the measure called Centimorgans) and that segment must exceed a set number of SNPs, the points that we measure. Of course, such a procedure is just a statistical test to estimate whether you are related. In reality, you might be and the procedure does not pick up that you are. What we consider a match is typically a half match because the match is only with one of the chromosome pair. The choice of these two parameters determines how likely a “real match” is versus the possibility of a false-positive event. In future blog posts, the question of whether the chances of false-positive matches are overstated will be examined.
Current genetic genealogy theory says that DNA is passed from one parent or the other to the child in long strands. The point at where a change occurs is called a recombination point or crossover. These recombination events are believed to be rather infrequent. One source referenced in the Recombination page on the ISOGG Wiki says that males average 27 crossovers per child and females average 41. That would be over 22 chromosomes of varying length. Sometimes a whole chromosome is thought to be passed down intact. Usually that involves one of the shorter, high numbered chromosomes.
Interestingly, we think of these long strands of DNA as either belonging to the paternal side or the maternal side of the chromosome pair, but the individual point on the strand called an allele cannot be determined by our testing technology as to which side that it is on. I believe what we have here is a case of people thinking that they see what they don’t see. For various reasons, I have developed a contrarian view about DNA being passed in long segments. As you see from my chain letter analogy, I believe that a person typically has several lines back to a more distant common ancestors and the DNA that manages to survive coming down the different lines reconstructs that segment of the ancestor’ DNA corresponding to the match. Another point to consider is that comparing a specific SNP between two people, you have about a 75 percent chance of having at least a half match at random. Just these random matches can be the “glue” that holds many small matches together to give us the appearance of long strands being passed. I have more arguments to support this belief, but I will save them for later blog posts.
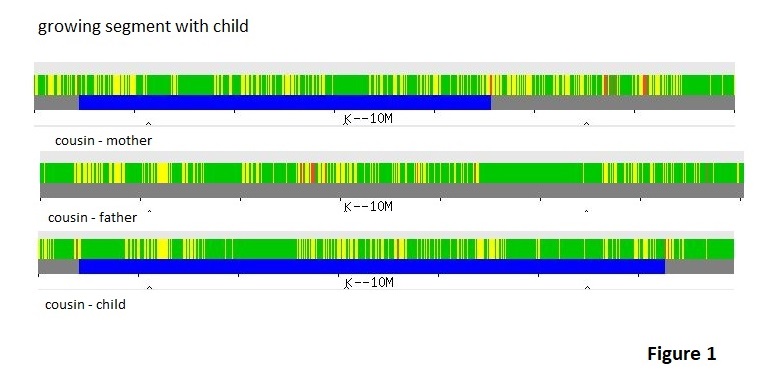
In my view, I see that switching between parents’ contributions is so common that the idea of infrequent crossovers is a faulty part of the theory. Perhaps I can illustrate this with an example. In Figure 1 we have a couple of GEDmatch Chromosome Browser images for ‘one-to-one’ comparisons on chromosome #20. The top comparison is between a child and her matrilineal grandmother. The graphic shows mostly yellow for half matches with small bands of green for full matches. The blue line confirms that the matching segment is believed to be the whole chromosome.
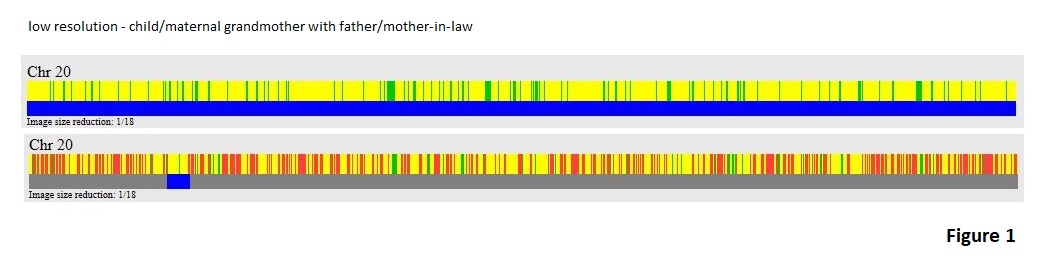
The lower comparison is between the father of the child and the father’s mother-in-law. Of course, the mother-in-law is the same person as the matrilineal grandmother in the first comparison. A One-to-one comparison between the father and the mother on GEDmatch using default parameters does not estimate a relationship nor does the father/mother-in-law comparison, so the father and mother are not likely related in the last couple of hundred years. Returning to the lower display in figure 1, we still have a lot of yellow with generally fewer and less broad bands of green with a lot of red areas mixed to signify no match areas. I used parameters of 300 SNPs and 3.0 cM minimum segment size for the comparison rather than defaults parameters of 500 and 7.0 cM so that I could get a small matching segment to show. Some will say that the matching segment could be a false-positive since I used such small segment size, but in an upcoming blog post I will show that the chances of getting a false-positive comparing two Family Finder kits using those parameters is negligible. For now, just trust me about that.
At first glance, the match between the child and grandmother appears to be just the grandmother’s entire recombined chromosome #20 contribution being passed down as expected to her daughter, the child’s mother, and that chromosome survived intact the recombination process of the parents passing their DNA to the child. The small matching segment between the father and his mother-in-law is in an area that is generally yellow in both comparisons indicating a half match. The conventional wisdom conclusion is that the match between the father and mother-in law would be on the other chromosome in the pair.
I think the Chromosome Browser is misleading us here by the washing out many of the full match indications with the low resolution image. In Figure 2, we are focusing on the region where we showed a match between the father and the mother-in-law by using the parameters of 300 SNPs and 3.0 cM minimum segment length . Full resolution was selected, though I do not think it really is full resolution in the sense of one SNP equaling one pixel. None the less, the image has a lot more green indicating full matches and some areas of the green in the comparison between the father and his mother-in-law line up with those in the child/grandmother comparison. From this perspective, the father does seem to have some common ancestry with the mother-in-law and the father is adding some DNA to the match between the child and the maternal grandmother. You cannot make the conclusion that the whole chromosome has been passed down from the grandmother, through the child’s mother, and then to the child after seeing it this way.
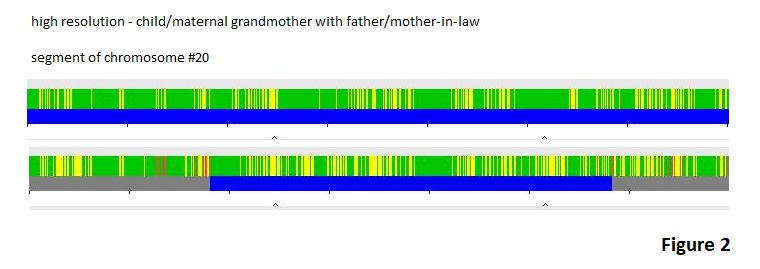
This is not a rare example. You easily can find examples like this if you just look at the data. In the next few blog posts I will present more evidence that the idea of long strands of DNA being passed from one or the other parent during recombination is incorrect and that crossovers are far from being infrequent.
Upcoming attraction: The Misunderstood Short Segment Match